Services
iSilico offers advanced computational tools to streamline and enhance the drug discovery process, empowering scientific research for greater efficiency and precision.

Our Expertise
We specialize in advanced computational tools and analysis, supporting detailed molecular research and accelerating discoveries in drug development and biological sciences
Drug Discovery
Utilized Software
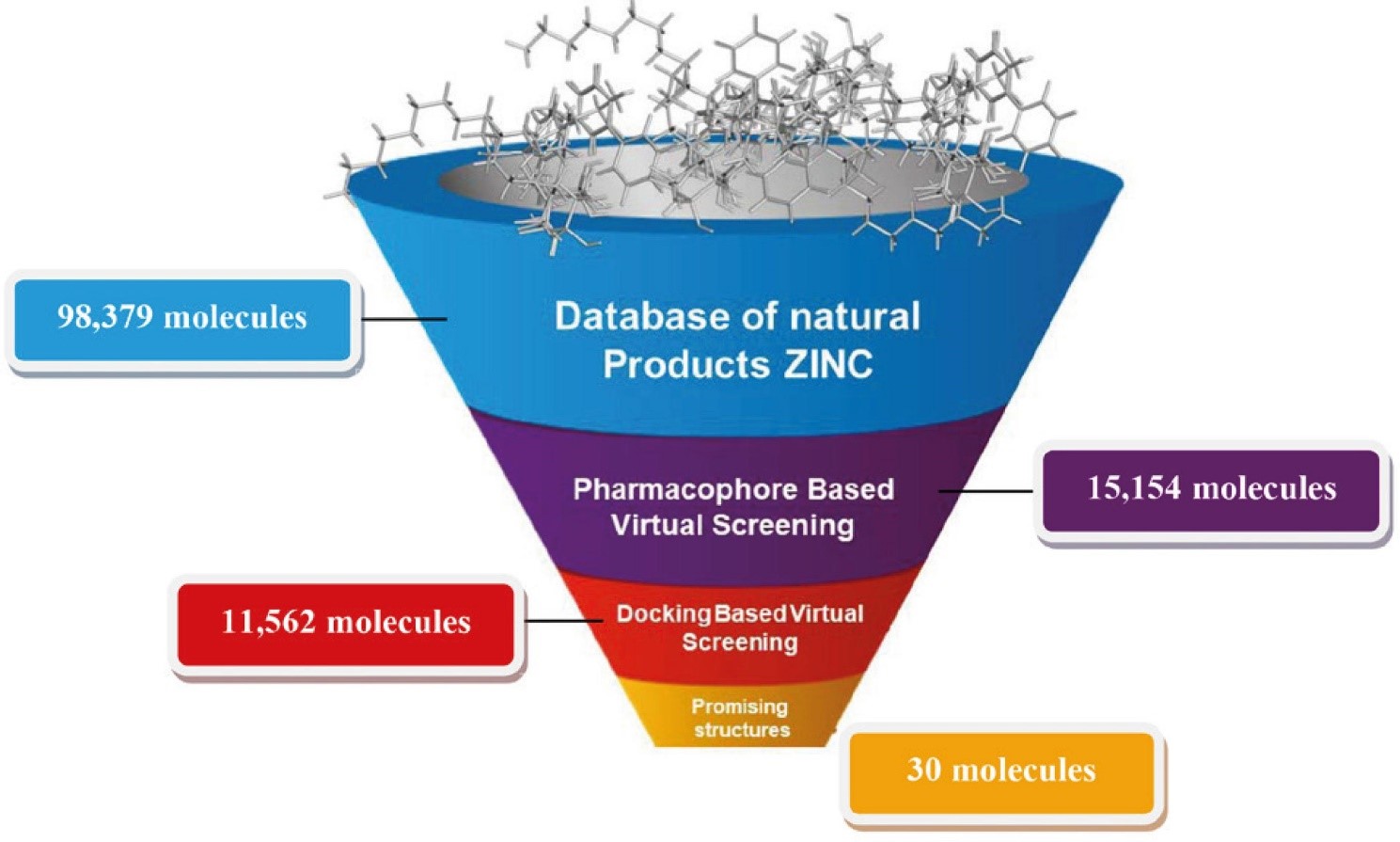
Database Mining
Virtual screening can be defined as a set of computational methods that analyzes large databases or collections of compounds in order to identify potential hit candidates.
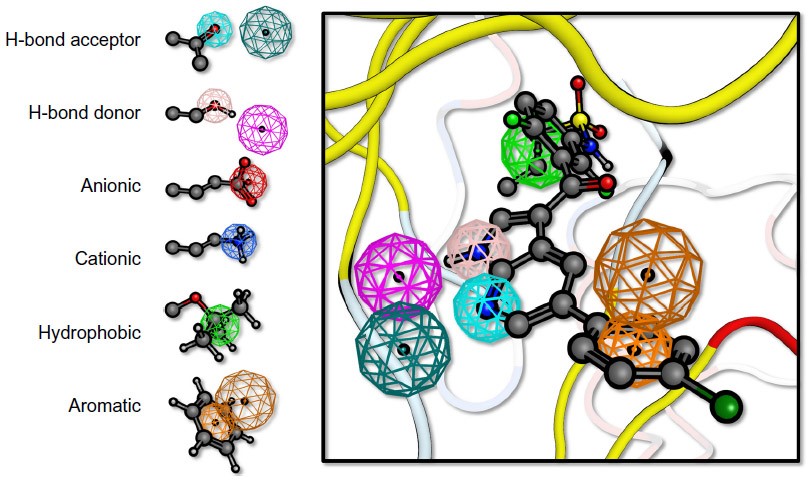
Pharmacophore Modeling
Pharmacophore modeling detects essential ligand-receptor interactions, focusing on steric and electronic features, with stages for dataset preparation.

Molecular Docking
Molecular docking evaluates molecule fit in target sites, using algorithms to create poses ranked by scoring functions for optimal binding orientation.
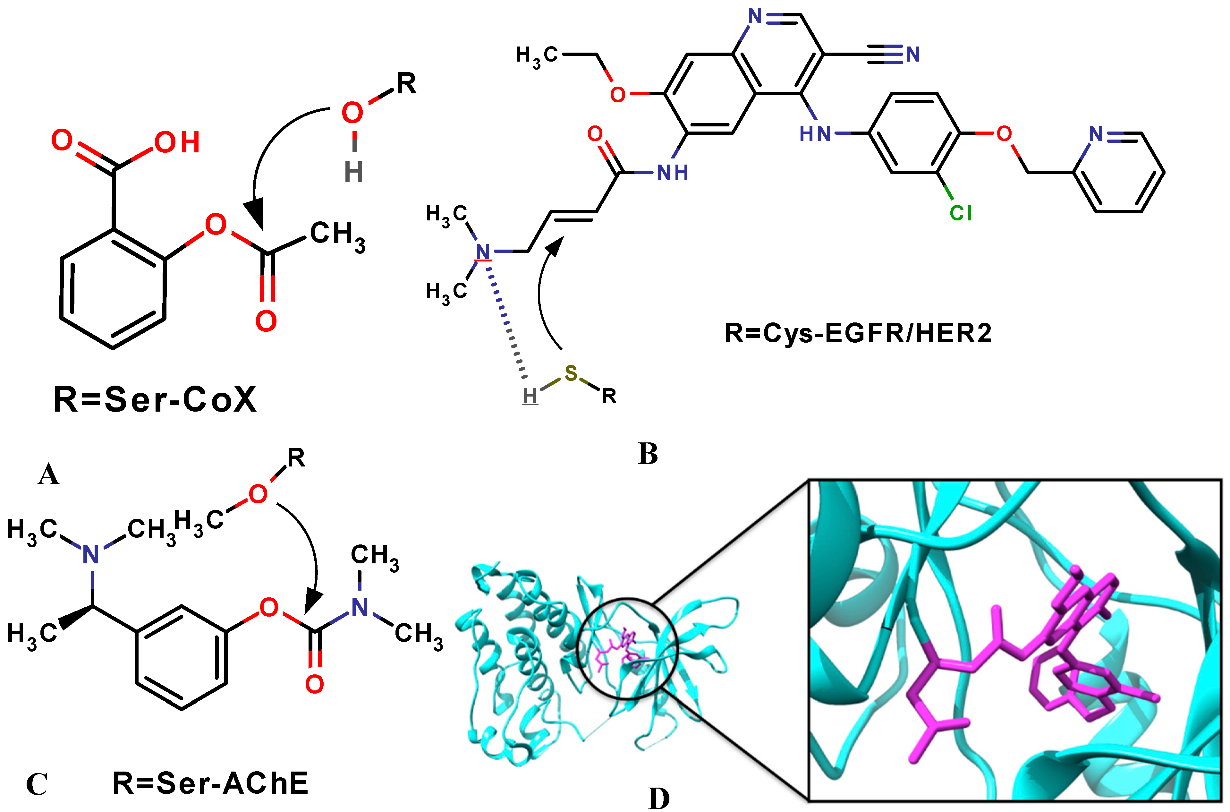
Covalent Molecular Docking
Covalent ligands, i.e., compounds that irreversibly bind to the receptor and possess a reactive moiety to target specific amino acid residues, helped to solve some of these obstacles.
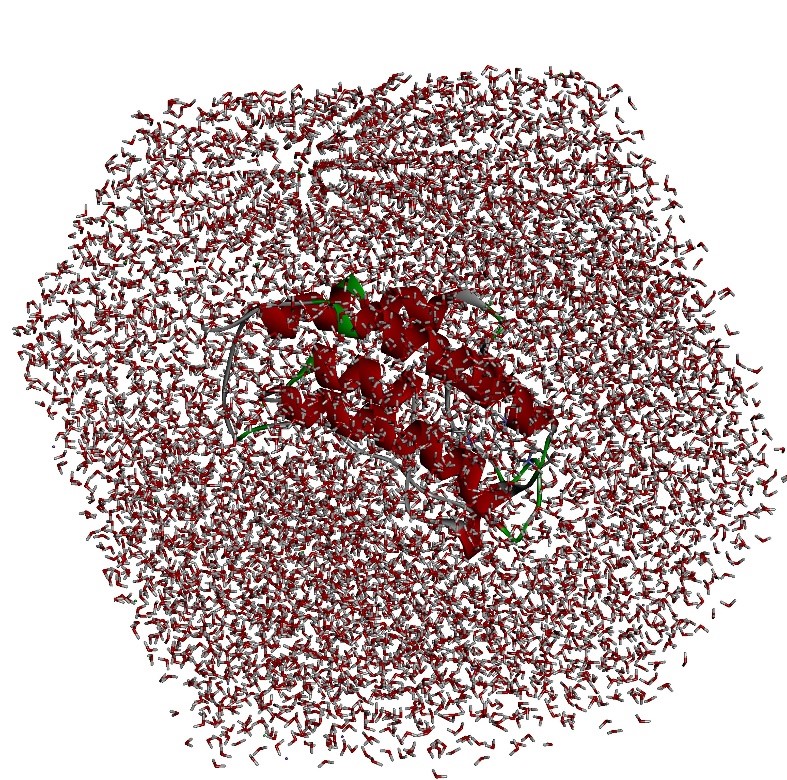
Molecular Dynamic Simulations
Molecular dynamics simulations analyze atomic and molecular interactions, solvation, and conformational changes of proteins and compounds under different conditions.
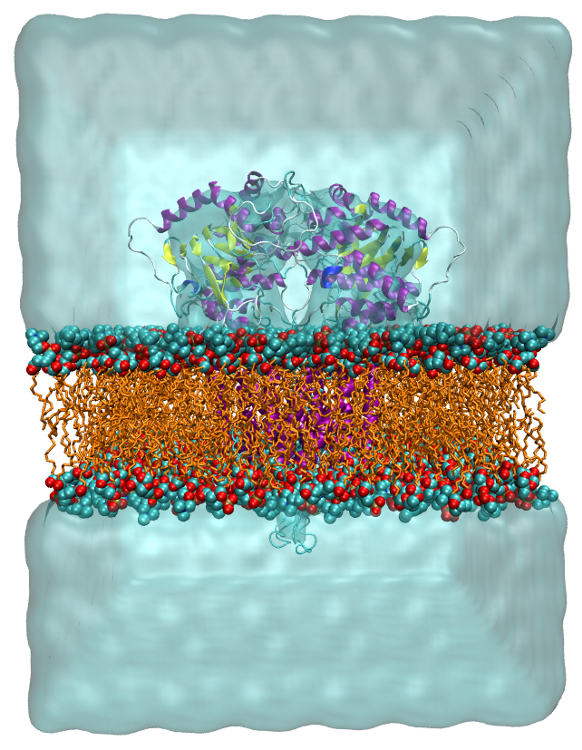
Protein-Membrane Simulations
Simulating membrane proteins in realistic environments is crucial for understanding their structure, physiological roles, and involvement in diseases, despite study challenges.
Prices
Materials Science
Utilized Software
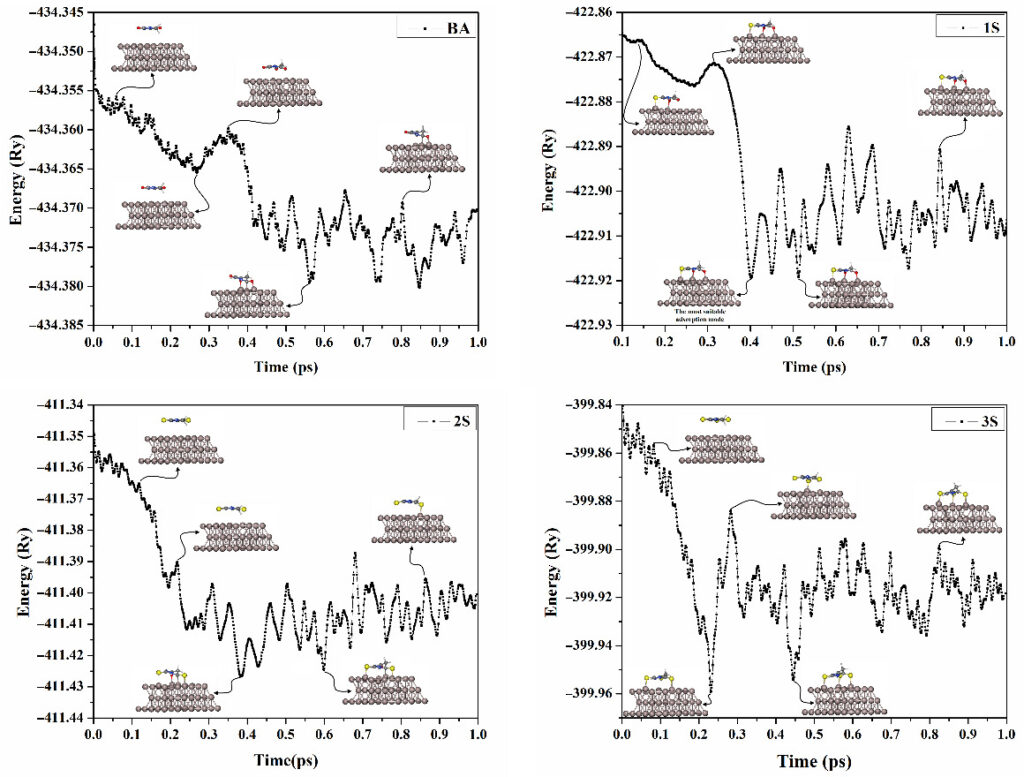
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations are announced as an appropriate method for determining the adsorption behavior of the adsorbates on substrates.
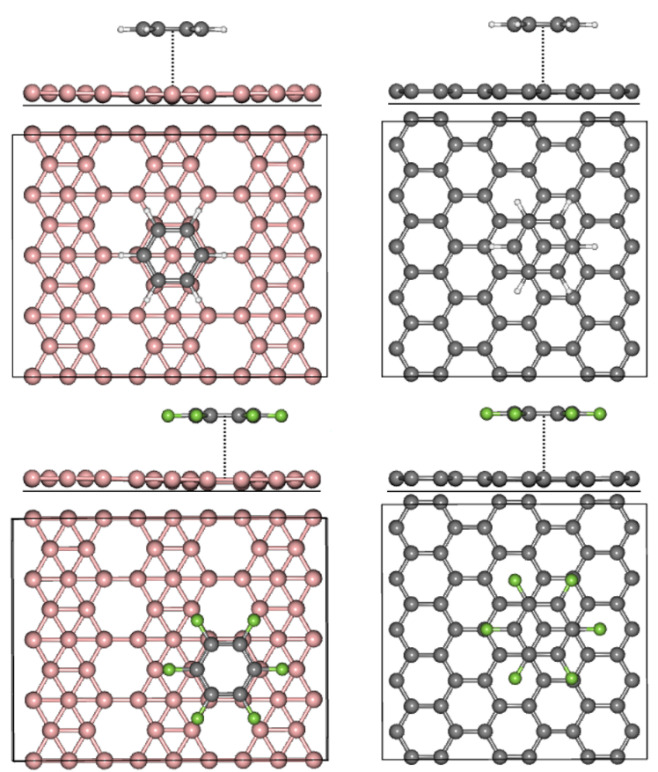
Adsorption Process
Towards a better understanding of the adsorption process within the adsorbate∙∙∙substrate complexes, adsorption energies will be computed at the equilibrium distances.

Bader Charge Method
Bader charge analysis provides insights into charge transfer during adsorption, generating charge density difference maps for adsorbate-substrate complexes visualized via VESTA.
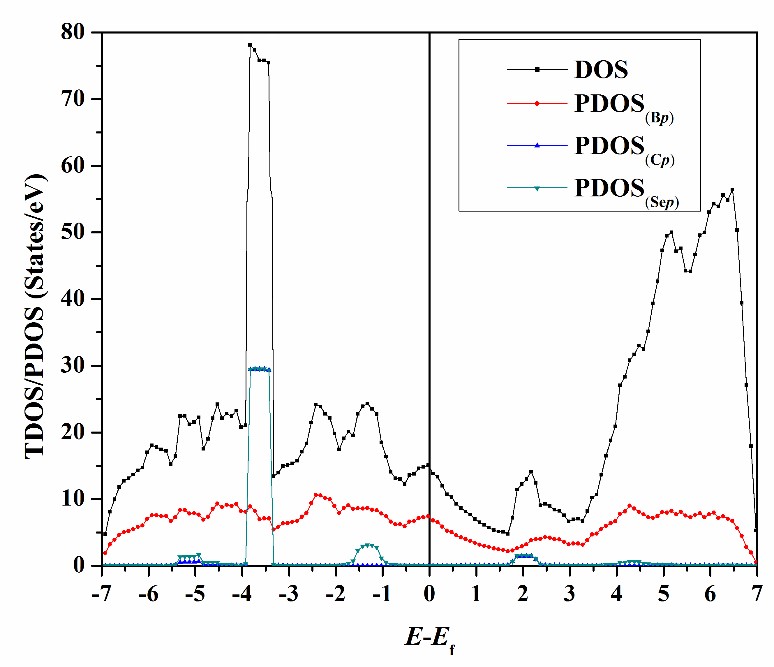
Density of States (DOS)
TDOS and PDOS analyses reveal interaction nature in adsorbate-substrate complexes, highlighting each orbital’s role in the adsorption process.
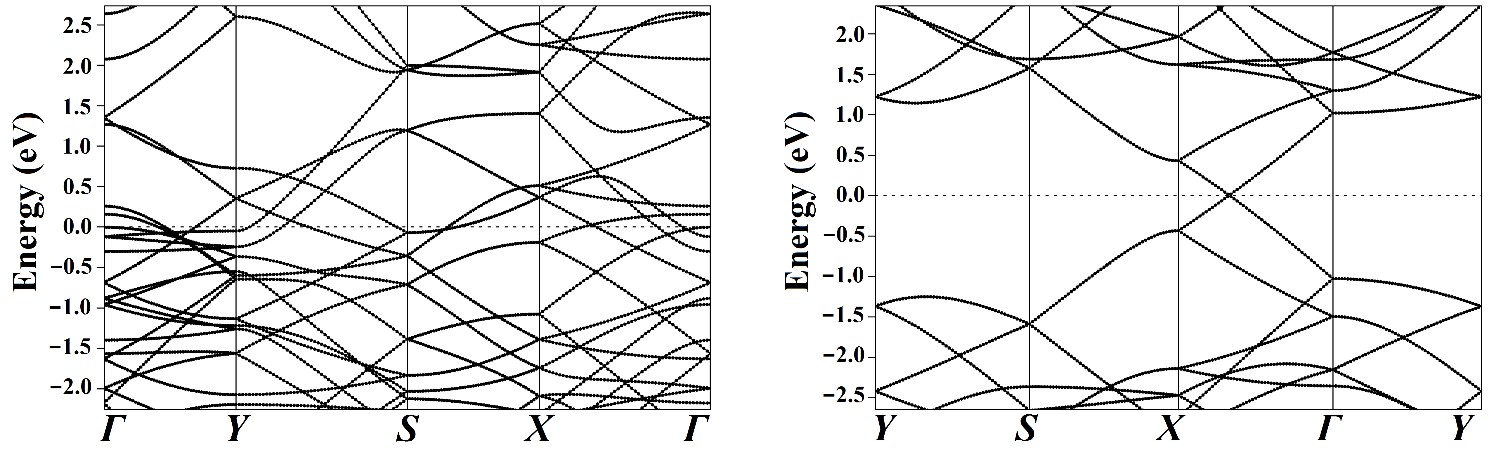
Band Structure
The electronic band structure analyses were performed to illuminate further the effects of the adsorbents on the electronic properties of the adsorbate.
Prices
Quantum Mechanical Calculations
Optimization of Complexes Using Gaussian09 Software
Optimizing molecular complexes through Gaussian09 software for accurate structural and energy analysis
Interaction Energy Calculations
Calculating interaction energies to assess the strength and nature of molecular interactions within complexes

Quantum Theory of Atoms in Molecules (QTAIM)
Applying the Quantum Theory of Atoms in Molecules (QTAIM) to analyze electronic properties and molecular interactions at the atomic level.

Noncovalent Interaction (NCI) Index
The NCI index, using electron density and its derivatives, reveals intermolecular interaction origins through 3D NCI plots generated via Multiwfn and VMD software.
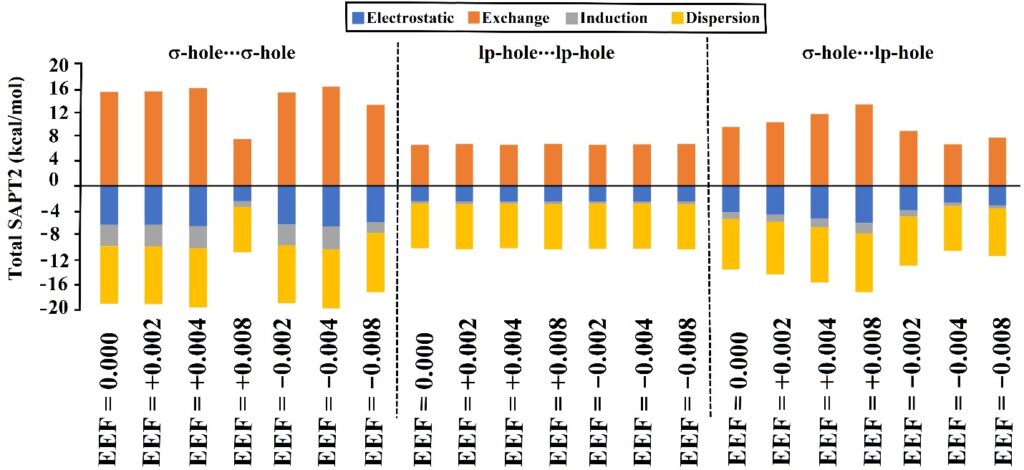
SAPT-EDA
SAPT-EDA is executed to elucidate the physical nature of noncovalent interactions. Utilized software: PSI4 code.
Prices
Drug Delivery
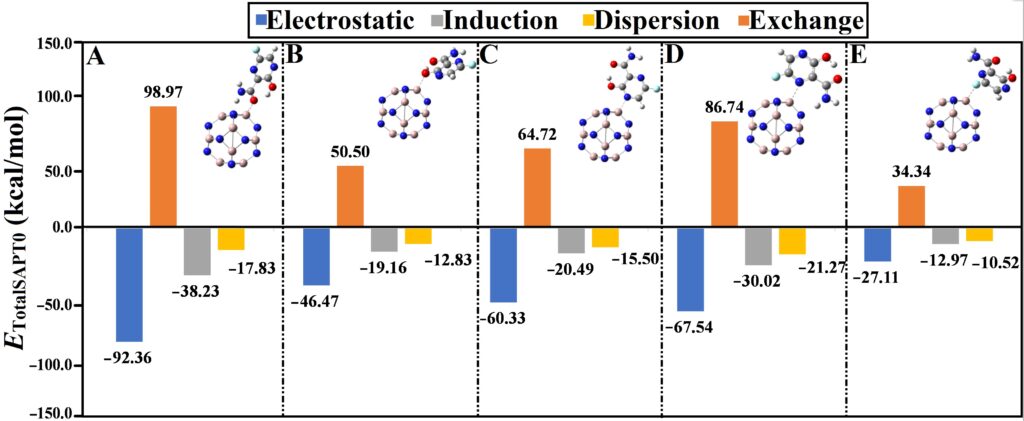
SAPT
SAPT analysis decomposes total adsorption energy into electrostatic, induction, dispersion, and exchange components, providing interaction insights via the PSI4 package.
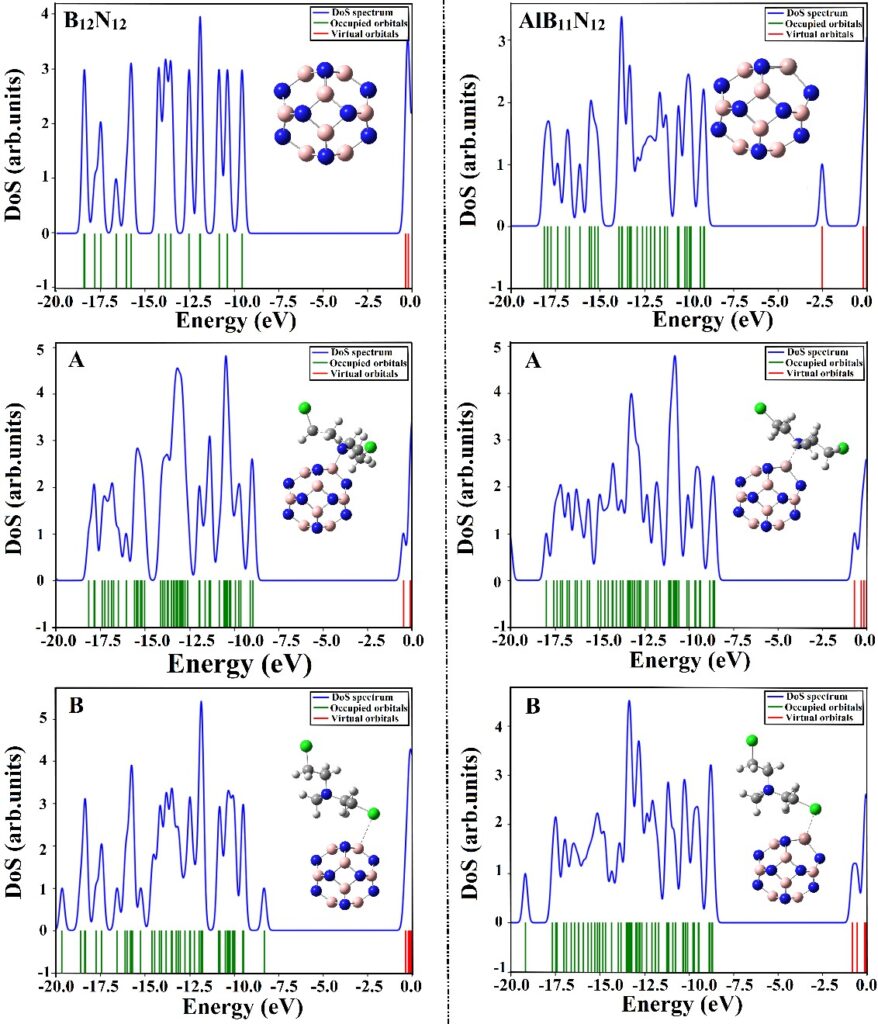
Density of state (DOS)
DOS diagrams reveal electronic property changes before and after adsorption, highlighting shifts in HOMO, LUMO, and Fermi levels using GaussSum software.
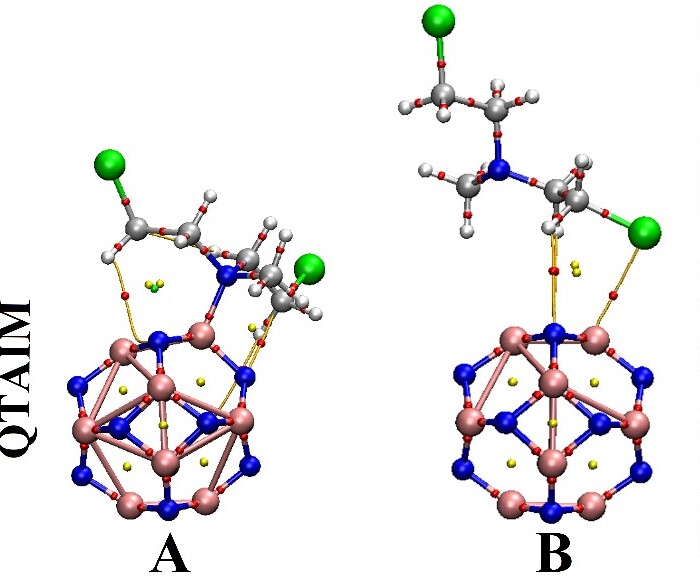
QTAIM
QTAIM analysis uses bond paths, critical points, and topological features to study drug-nanocarrier adsorption, using Multiwfn and VMD
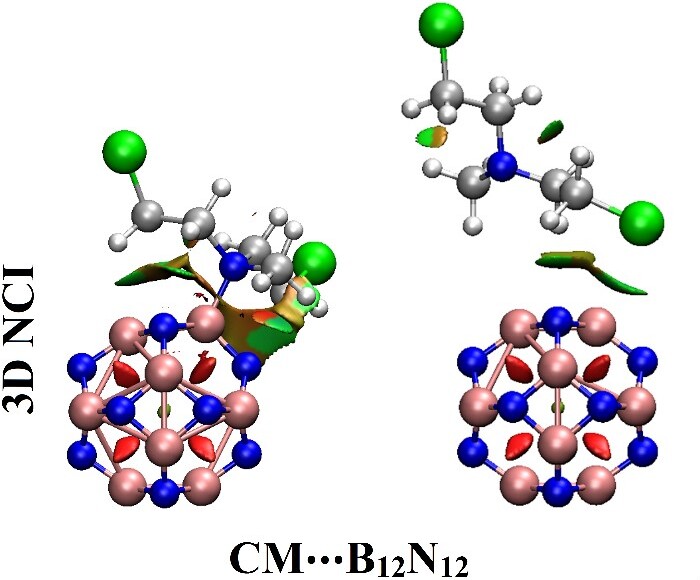
Noncovalent interaction (NCI) index
NCI index analysis visualizes inter- and intra-molecular interactions in adsorption, using 3D iso-surfaces colored by sign(λ2)ρ, with Multiwfn and VMD software
Prices
Premium Scientific Research Services
We assist with data analysis and manuscript preparation for high-quality research,Our team offers tailored insights for effective navigation of scientific challenges.
